Section: New Results
Computational Physiology
Tumor Growth Modeling
Participants : Erin Stretton [Correspondant] , Nicholas Ayache, Hervé Delingette, Bjoern Menze, Ender Konukoglu, Ezequiel Geremia, Emmanuel Mandonnet.
This work was funded by Care4me.
-
Performing a sensitivity analysis of long time series of multi-modal data.
-
Improving our current models and their inputs, including creating a white matter and brain mask template with the help of a neurosurgeon and integrating the Powell bound constrained optimization into the minimization routine.
-
Performed comparison of using a patient DTI, an atlas DTI or no DTI at all showing the difference in accuracies in the simulation results since the patient DTI is not always available in a clinical setting or is of poor quality. We found that using an atlas DTI produced only slightly less accurate results than using a patient DTI, where as using no DTI at all did not produce accurate results.
Synthetic Echocardiographic Image Sequences for Cardiac Inverse Electro-Kinematic Learning
Participants : Adityo Prakosa [Correspondant] , Maxime Sermesant, Hervé Delingette, Eric Saloux [CHU Caen] , Pascal Allain [Philips Healthcare] , Pascal Cathier [Philips Healthcare] , Patrick Etyngier [Philips Healthcare] , Nicolas Villain [Philips Healthcare] , Nicholas Ayache.
This work is done in collaboration with Medisys, Philips Healthcare Suresnes, France, and Cardiology Department of CHU Caen, France.
-
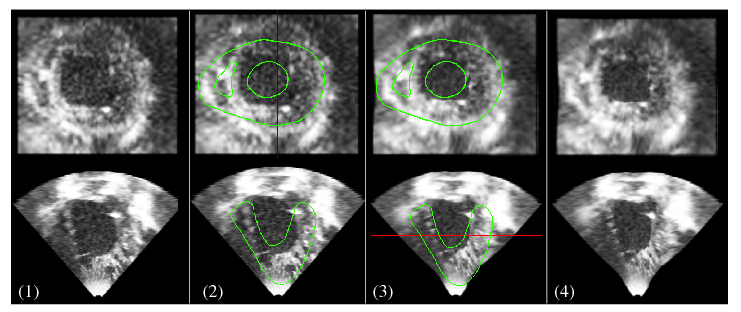
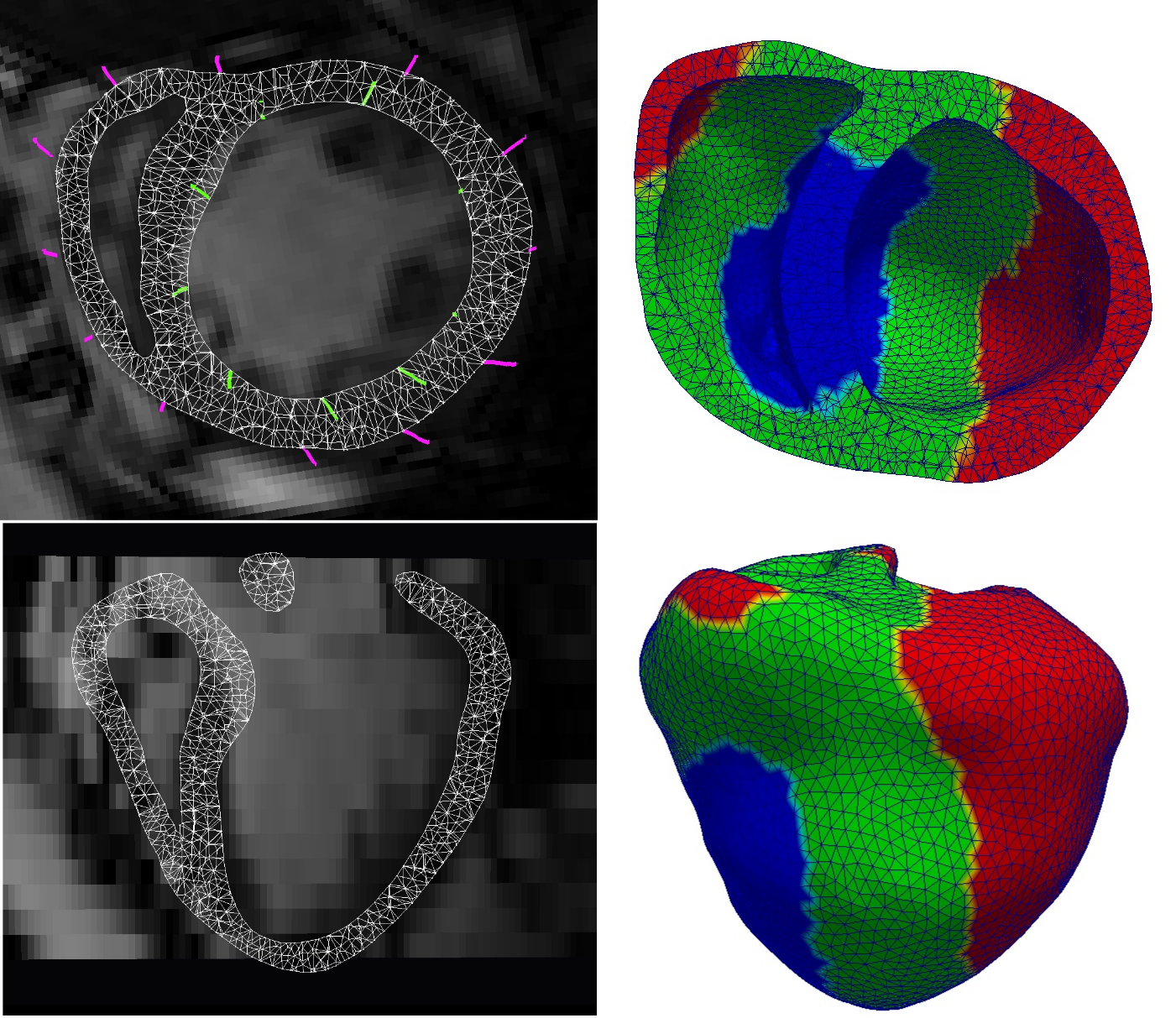
A database of 120 synthetic 3D echocardiography (US) sequences is created based on a cardiac electromechanical model (see Figure 13 ).
-
Kinematic descriptors are extracted from the displacement field estimated from the synthetic 3D US sequence using the iLogDemons non-rigid registration method [37] .
-
Cardiac inverse electro-kinematic learning is done by using the database of synthetic 3D US sequences in order to estimate the cardiac depolarization times for the given kinematic descriptors [67] . First evaluation on two clinical sequences from patients with Left Bundle Branch Block shows encouraging results.
|
Prediction of patient-specific Ventricular Tachycardia for radio-frequency ablation therapy planning
Participants : Jatin Relan [Correspondant] , Maxime Sermesant, Hervé Delingette, Nicholas Ayache.
This work is funded by the FP7 European Project euHeart.
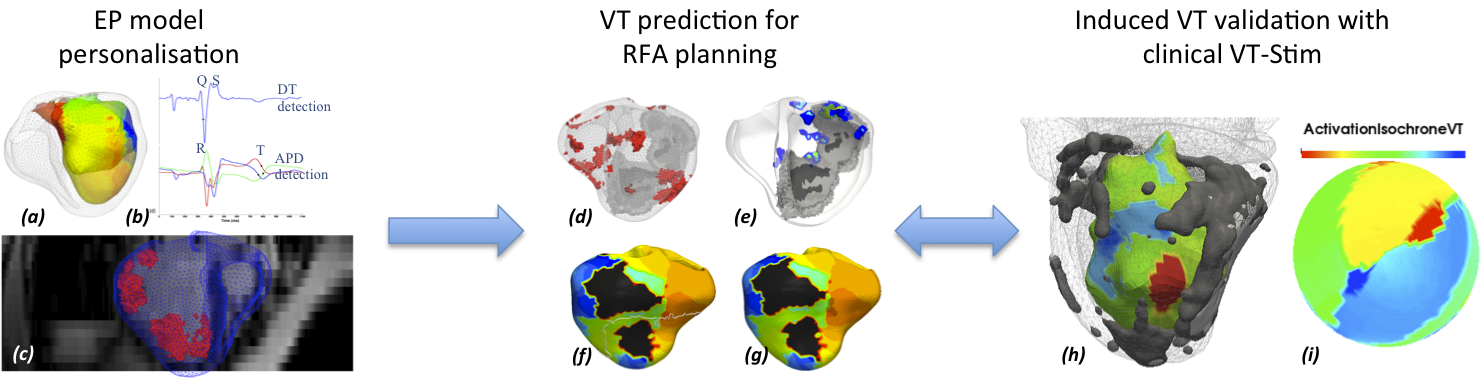
In this work, we build a patient-specific cardiac electrophysiology (EP) model derived from hybrid XMR imaging and non-contact electro-anatomical mapping procedure on a patient with heart failure. The model is then used to predict patient-specific arrhythmias, such as induced ischemic Ventricular Tachycardia (VT) (Fig. 14 ) and leads in generation and evaluation of patient-specific VT circuits, with critical exit points for Radio Frequency (RF) ablation. These predictions are now validated with some clinical cases, with electrophysiology mapping of induced VT in patients undergoing the clinical VT-Stim procedures (Fig. 14 ).
|
Real-time simulation of catheter ablation in the context of cardiac arrhythmia
Participants : Hugo Talbot [Correspondant] , Federico Spadoni, Maxime Sermesant, Hervé Delingette, Stephane Cotin.
This work is performed in the context of the euHeart project and the PhD of Hugo Talbot in collaboration with the Shacra (INRIA, Lille Nord Europe) team.
-
This work aims at simulating in real-time the endovascular procedure of radiofrequency ablation of the left ventricle for patient suffering from Ventricular Tachycardia.
-
Fast simulation of electrophysiology has been reached with a Eikonal model[40] .
-
Use the SOFA platform for simulating endovascular navigation and cardiac electrophysiology.
|
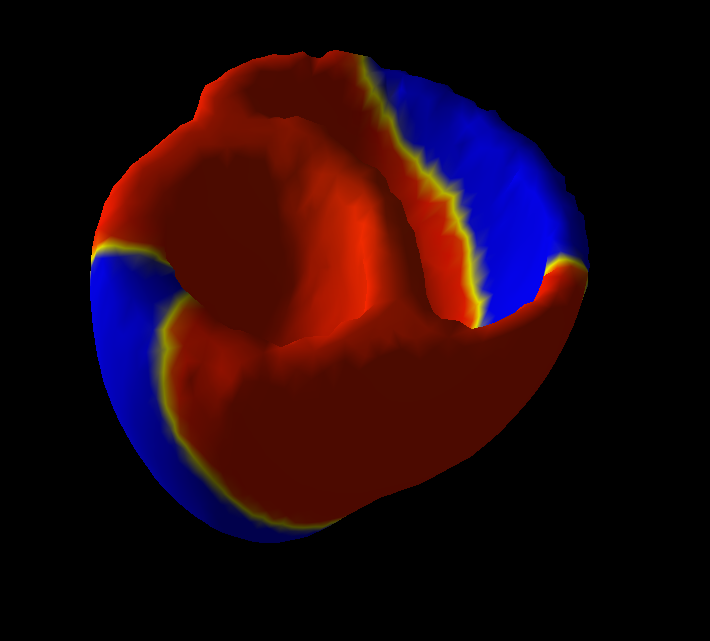
Personalized model of the heart for cardiac therapy planning
Participants : Stéphanie Marchesseau [Correspondant] , Ken C.L. Wong, Hervé Delingette, Maxime Sermesant, Nicholas Ayache.
This work has been performed in the context of the euHeart european project.
-
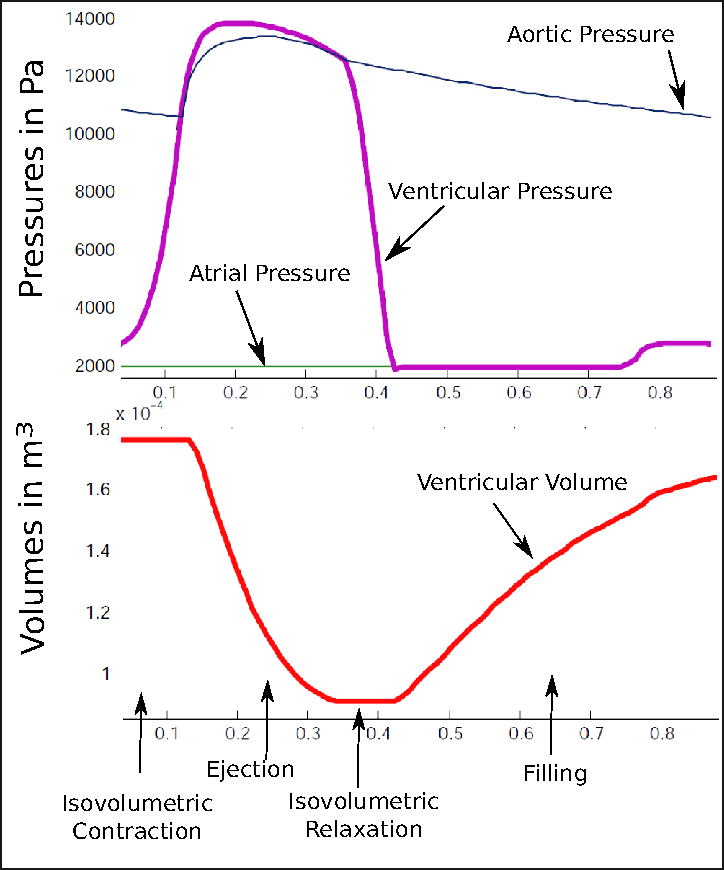
We improved the existing electromechanical model of the heart (see Fig 16 ) to include mechanical non linearity, viscosity and strain rate dependent contractility. It was implemented in SOFA with a new four valves model to deal with the cardiac phases and enforce isovolumetric constraint.
-
We have obtained first personalization of cardiac mechanics from 3 cine-MRI cases using a variational approach (adjoint method)[54] .